From FASTQ to Inside: All on one platform.
DNA Metabarcoding is powerful, but managing pipelines, versions, and data at scale can be challenging. Labs naturally focus on scientific questions and wet-lab excellence, not on building high-throughput, production-ready bioinformatics. spcfy provides a unified platform with transparent, versioned Illumina workflows and structured data handling, so you can focus on interpreting biodiversity data rather than maintaining infrastructure.
The Operational Bottleneck
The current challenges in DNA metabarcoding are technical, not conceptual. Fragmented workflows, custom maintenance scripts, and disconnected metadata do not question the validity of the science itself, but they drastically reduce efficiency and reproducibility. spcfy addresses this infrastructure layer directly: we provide a unified environment for data handling, automated QC, and transparent pipelines, allowing you to focus purely on interpretation.
Technical hurdles that consume your time:
Inconsistent Protocols
Conflicting primer sets and reference DBs often limit cross-study comparability.
Metadata Chaos
Disconnected spreadsheets and missing fields block meaningful data integration.
Undocumented QC
Manual steps and unversioned pipelines put your reproducibility at risk.
Manual Scripting
Writing custom scripts for every plot consumes valuable research time.
One upload. One workflow. One platform.
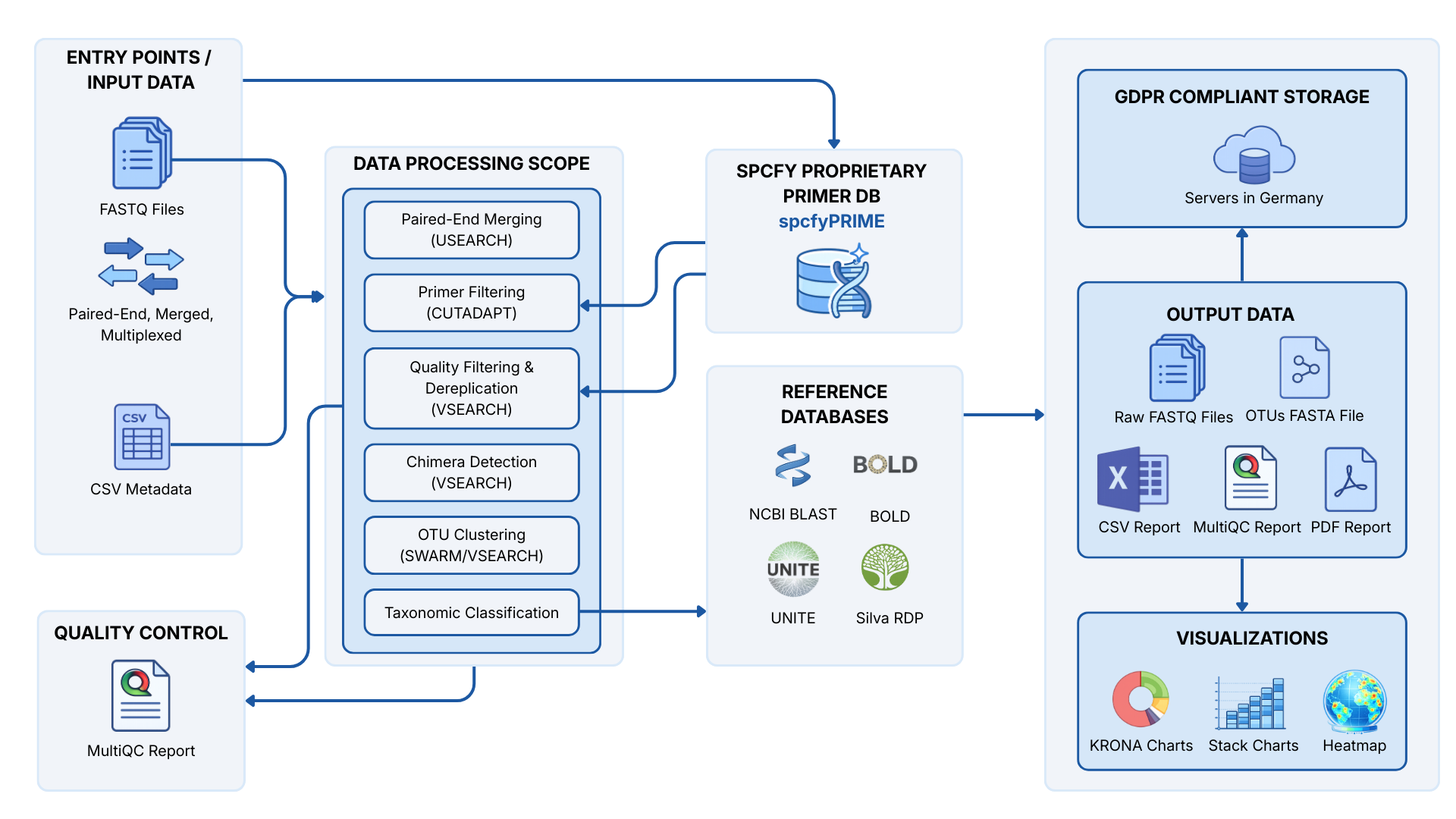
spcfy is designed around your Metabarcoding workflow: Uploading FASTQ files, structuring metadata, processing OTUs, assigning taxonomy, and interpreting results across projects. By integrating all relevant steps of DNA Metabarcoding analysis, we want to help bioinformaticians and laboratories standardize repetitive tasks, thereby enabling them to refocus on the scientific part of their work.
Raw Data Ingestion & Automated QC
Upload raw Illumina FASTQ files (single or paired-end) via drag-and-drop. spcfy immediately performs automated quality control: checking Phred scores, removing primers/adapters, and executing trimming operations before data enters the pipeline with no manual scripting required.
Standardized Amplicon Workflows
Select from pre-configured, versioned pipelines for 16S, CO1, 12S, ITS2, or PITS2. Our workflows utilize established algorithms (based on VSEARCH/SWARM logic) for denoising, chimera removal, and clustering into OTUs. Every analysis is fully documented and reproducible, including all parameter settings.
Curated Taxonomic Assignment
Rely on maintained reference databases instead of local flat files. We integrate and update standards like SILVA (bacteria), MIDORI (metazoa), and UNITE (fungi). The system automatically selects the appropriate references for your marker choice and assigns taxonomies with calculated confidence intervals.
Dynamic Metadata Integration & Stats
Link sequence data directly to your sample sheet (GPS, habitat, time series, environmental factors). Calculate alpha and beta diversity (Shannon, Simpson, Bray-Curtis) on the fly. Our interactive tools allow you to dynamically filter and group PCoA plots and heatmaps by metadata, without writing a single line of R code.
Full Data Sovereignty & Compliance
spcfy is not a black box. You retain full control over your data. Export results at any time in standard formats (CSV, FASTA) for further processing in Phyloseq or Vegan. All data is hosted in Germany and processed according to strict security standards.